Loading EEGLAB .set files in R - part 1
Like a lot of people, I’ve been using EEGLAB and Fieldtrip for years and have a lot of data already processed using those packages. It can be a bit annoying getting the data from them - in the past I’ve converted the data to text/csv files, which is ok as far as it goes. It’s a bit of a faff getting them in the right format, and EEGLAB’s in-built export function drops useful info like epoch numbers and event codes etc. Fortunately for us, there is a way to load Matlab files directly in R using the R.matlab package. Fieldtrip has no native saving function, so you simply save .mat files directly, while EEGLAB .set files are also just Matlab .mat files with a different.
That said, there a couple of things to keep in mind in order to smoothly load the data in. Matlab file formats have changed over the years, and it’s easier to get some things out of the earlier formats than the recent, HDF5 format. I’ll start off showing what happens with HDF5 files, thanks to the h5 package. In the follow-up post, I’ll try again with R.matlab and older file formats.
library(h5)EEGLAB .set files - HDF5-style
There are a couple of important options in EEGLAB that determine whether the file format used is Matlab v6.5 or v7.3, and whether the data is saved as a single .set file, which contains the data and all the associated EEG structure, or as two files, with the EEG structure saved in a .set file and the actual data saved in an .fdt file.
I usually use the dual-file format anyway, as that results in smaller files.
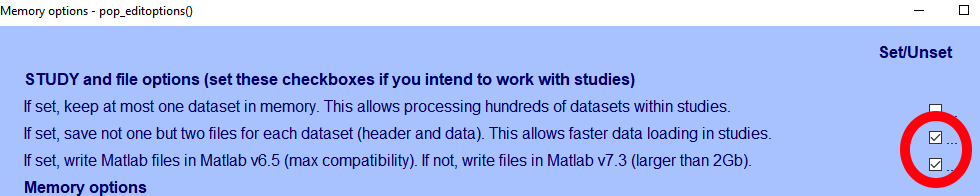
(#fig:eeglab_memo)The two critical options!
Despite the ticks in the pic above, I’ve saved a file with both thos options unticked - a v7.3 file with the data all in a single file. Let’s load it in using the h5 package.
file_name <- "HSFObj_hdf5.set"
eeg_data <- h5file(file_name)
eeg_data## H5File 'HSFObj_hdf5.set' (mode 'a')
## + #refs#
## + EEGh5close(eeg_data)HDF5 files have a hierarchical structure organised into Groups and Datasets. Our file has two groups - #refs# and EEG. We address the EEG group like so:
eeg_data <- h5file(file_name)
eeg_data["EEG"]## H5Group '/EEG'
## + chaninfo
## + chanlocs
## + epoch
## + etc
## + event
## + reject
## + stats
## + urevent
## D comments
## D condition
## D data
## D datfile
## D dipfit
## D epochdescription
## D eventdescription
## D filename
## D filepath
## D group
## D history
## D icaact
## D icachansind
## D icasphere
## D icasplinefile
## D icaweights
## D icawinv
## D nbchan
## D pnts
## D ref
## D saved
## D session
## D setname
## D specdata
## D specicaact
## D splinefile
## D srate
## D subject
## D times
## D trials
## D urchanlocs
## D xmax
## D xmin
## A MATLAB_class
## A MATLAB_fieldsh5close(eeg_data)As you can see, the EEG group corresponds to the EEG structure that was saved from Matlab. There are a bunch of H5 groups ( marked with + ), datasets (D), and attributes (A) that we can try to access by adding their label to the root group (“EEG”) using “/whatever”:
eeg_data <- h5file(file_name)
eeg_data["EEG/data"]## DataSet 'data' (54 x 717 x 68)
## type: numeric
## chunksize: 54 x 4 x 68
## maxdim: UNLIMITED
## compression: H5Z_FILTER_DEFLATE
## Attributes:
## A H5PATH
## A MATLAB_classh5close(eeg_data)This just describes the contents of the dataset rather than actually giving you the contents - for that you need to got a little further. You can treat the datasets as like a vector or a matrix, and address them using standard R syntax, and a fun thing about HDF5 files is it’s easy just to read parts of the data if you want. For example, if you just wanted the first ten elements, you would address it using [1:10]. Here I’ll go ahead and get all the data - 54 trials, 717 time points, 68 channels. We’ll also grab the times dataset.
eeg_data <- h5file(file_name)
actual_data <- eeg_data["EEG/data"][]
times <- eeg_data["EEG/times"][]
h5close(eeg_data)
dim(actual_data)## [1] 54 717 68plot(times[170:400], actual_data[1, 170:400, 64], type = "l")
So we’ve got our EEG data and a vector of times, and we can now do whatever we like with it! But the EEG data is just a bare matrix. We don’t have electrode labels (so we don’t know which is which) and we don’t have any trigger information, so if we have different trial types, we won’t be able to tell which one is which. In EEGLAB, channel information is stored in the chanlocs element of the EEG structure, so we can try to get that here.
eeg_data <- h5file(file_name)
eeg_data["EEG/chanlocs"]## H5Group '/EEG/chanlocs'
## D labels
## D radius
## D ref
## D sph_phi
## D sph_radius
## D sph_theta
## D theta
## D type
## D urchan
## D X
## D Y
## D Z
## A H5PATH
## A MATLAB_class
## A MATLAB_fieldsh5close(eeg_data)Looks great - there are all the things we would want - labels, locations etc. Let’s try to grab the labels.
eeg_data <- h5file(file_name)
print(try(eeg_data["EEG/chanlocs/labels"]))## [1] "Error in GetDataSetType(dsetptr) : Datatype unknown.\n"
## attr(,"class")
## [1] "try-error"
## attr(,"condition")
## <Rcpp::exception in GetDataSetType(dsetptr): Datatype unknown.>h5close(eeg_data)Hmm, it fails. How about the X co-ordinates?
eeg_data <- h5file(file_name)
print(try(eeg_data["EEG/chanlocs/X"]))## [1] "Error in GetDataSetType(dsetptr) : Datatype unknown.\n"
## attr(,"class")
## [1] "try-error"
## attr(,"condition")
## <Rcpp::exception in GetDataSetType(dsetptr): Datatype unknown.>h5close(eeg_data)Fails too. It turns out we can’t get anything out of the chanlocs structure. For technical details, there’s an issue for this over at the h5 Github page. We currently can’t get anything out of any of the groups within the root “/EEG” group - so any of the elements marked with a + in the list. It may be possible at some point, but not yet.
So if you’re satisfied with just getting the data out, the recent Matlab HDF5 files are absolutely fine. You’ll just have to marry them up with appropriate channel labels etc manually.
In a follow-up post, I’ll look at how to get the data from the older, v6.5 file format.